Deriving Gaussian Process Factor Analysis
April 20, 2025
Course project, deriving Gaussian Process Factor Analysis (GPFA)
In Spring 2025, I took the Bayesian Statistics course and did a group project on deriving and implementing an existing method from scratch. We picked this method because it is quite popular. My awesome teammate Clay Smyth and I stared at the equations for quite a bit but magically, we did it!
A short introduction
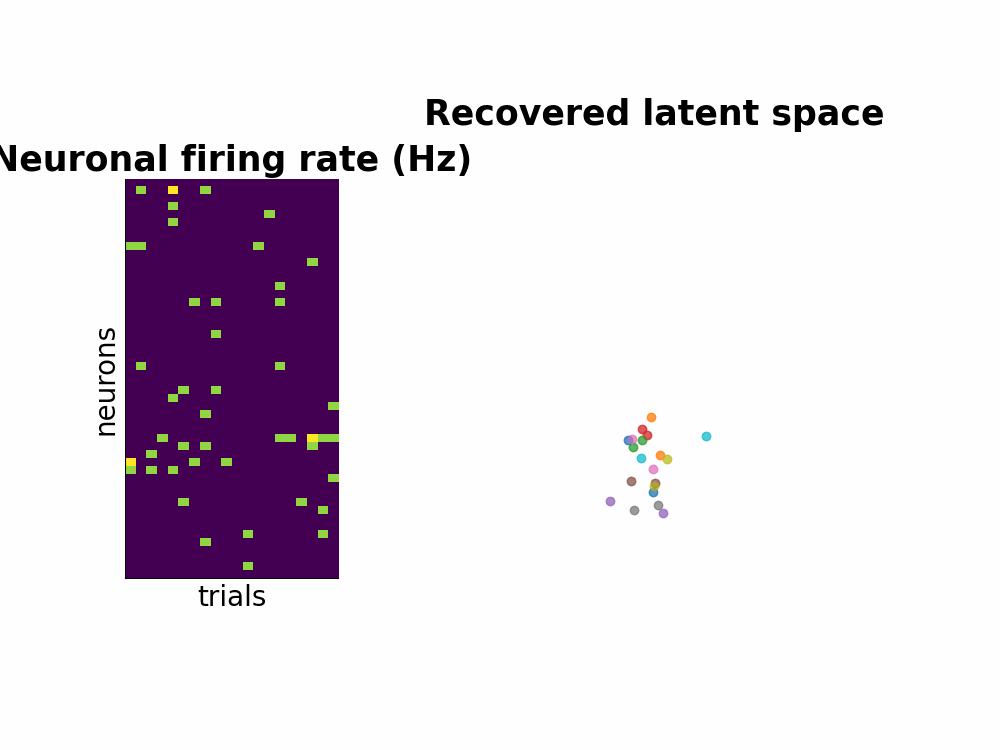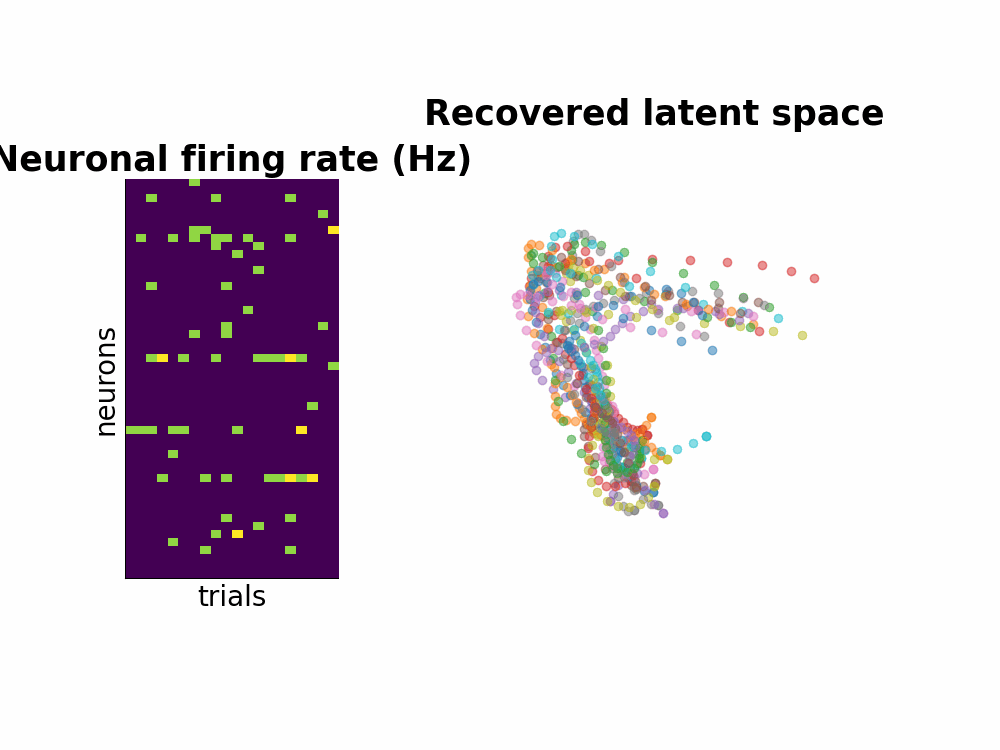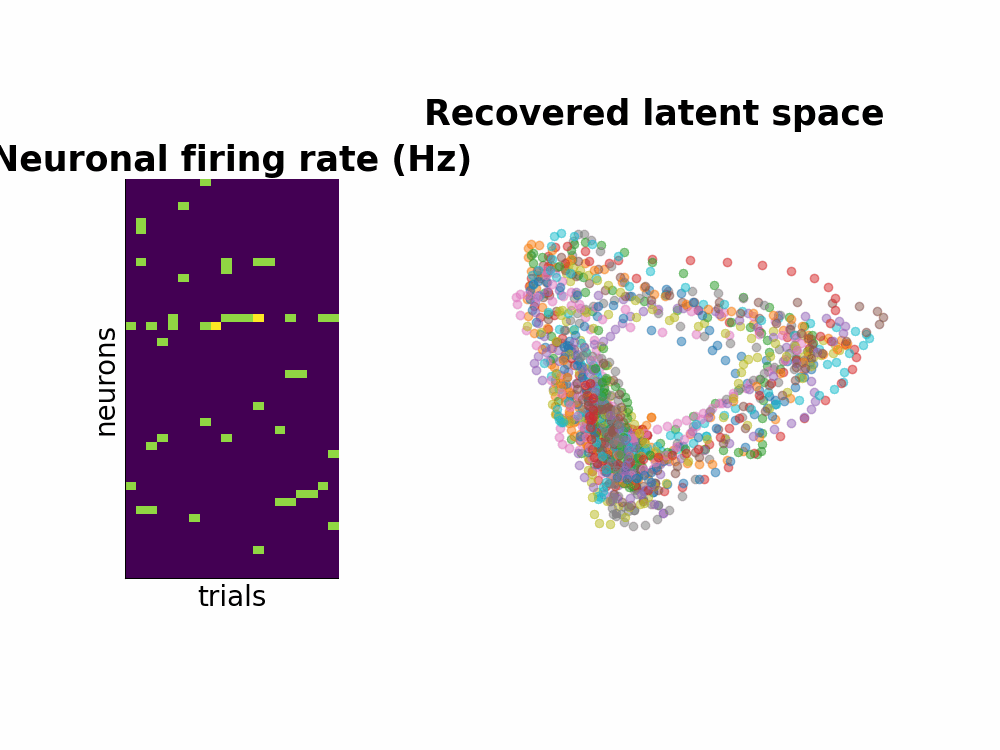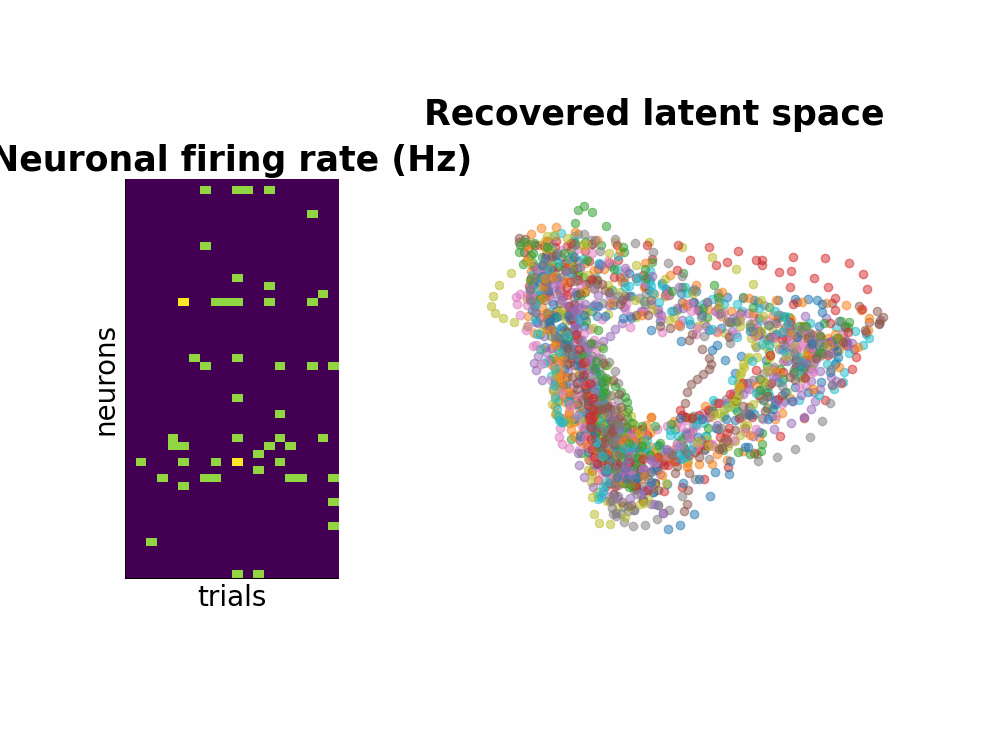
One way to study the brain is to directly measure the neuronal spiking activities. Spikes or also known as action potentials are binary sigals that neurons use to communicate with one another. Using a few electrodes, we can record hundreds or thousands of neurons spiking activities over seconds or minutes when the subject completes a task (for example, a decision making task or arm reach movements). This information conveyed by many neurons is a high-dimensional dataset. Neuroscientists usually attempt to understand this by smoothing the data and by dimensionality reduction techniques such as PCA.
Instead of seperating data smoothing and data dimensionality reduction into 2 steps, GPFA proposes a hierarchical model where the state variable is smooth - modeled by Gaussian process - and is related to observed noisy spikes through a linear regression. Every parameter is learned with Expectation Maximization method.
Here is the Github repo that contains all the walk through including the efficient implementation of matrix inversion.